I have two different estimates of an unknown variable with a known distribution. Each estimate has a different distribution of noise. I also have observations of a variable that depends on the same unknown variable.
I have tried to model it with a latent variable, but the samples I get seem to make no sense.
Can anyone please give me a hint of what is wrong with the model?
library(greta)
rm(list = ls())
#generation of latent variable (unavailable for the model)
real_shape=3
real_rate=0.01
latent_variable <- rgamma(10000, shape = real_shape, rate = real_rate)
#estimates (available)
variable1 <- latent_variable + rnorm(length(latent_variable), mean=10, sd=20)
variable2 <- latent_variable + rnorm(length(latent_variable), mean=20, sd=5)
#data depending on latent variable (available)
real_coef <- 1.5
data <-
latent_variable * latent_variable * real_coef +
rnorm(length(latent_variable), 0, 500)
#model variables
var1 <- as_data(variable1)
var2 <- as_data(variable2)
# transformed variables
dist_var1 <- fitdistrplus::fitdist(variable1[variable1 > 0], "gamma")
dist_var2 <- fitdistrplus::fitdist(variable2[variable2 > 0], "gamma")
#a priori distributions
#standard deviaton of the prediction error
sd <- cauchy(0, 10, truncation = c(0, Inf))
#estimate of the coefficient of the relationship between the latent variable and the observed variable
coef <- normal(0,10)
#estimate of the shape and rate of the distribution of the latent variable from the available data
shape <-
normal(
0.5 * (
dist_var1$estimate[["shape"]] + dist_var2$estimate[["shape"]]
),
dist_var1$sd[["shape"]] + dist_var2$sd[["shape"]]
)
rate <-
normal(
0.5 * (
dist_var1$estimate[["rate"]] + dist_var2$estimate[["rate"]]
),
dist_var1$sd[["rate"]] + dist_var2$sd[["rate"]]
)
#latent variable
latent_variable_estimate <- gamma(shape = shape, rate = rate, dim = length(latent_variable))
#standard deviations of the noise of the two estimates of the latent variable
sd_var1 <- cauchy(0, 0.5, truncation = c(0, Inf))
sd_var2 <- cauchy(0, 0.5, truncation = c(0, Inf))
#relationship between the estimates and the latent variable
distribution(var1) <- normal(latent_variable_estimate, sd_var1)
distribution(var2) <- normal(latent_variable_estimate, sd_var2)
#prediction
predict_data <- function(values, k)
{
prediccion <- k * values * values
}
prediction <- predict_data(values = latent_variable_estimate, k = coef)
distribution(data) <- normal(prediction, sd)
#model
m <- model(sd, coef, shape, rate, sd_var1, sd_var2)
#samples
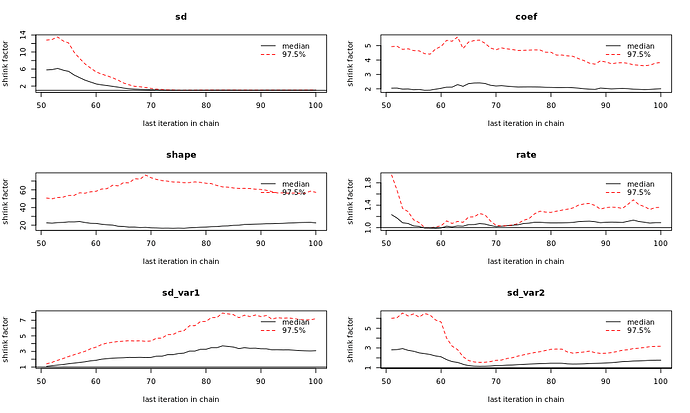
samples <- mcmc(m, n_samples = 100, warmup=1000, n_cores=2, chains = 2,one_by_one = TRUE)
summary(samples)
I’m getting:
summary(samples)
Iterations = 1:100
Thinning interval = 1
Number of chains = 2
Sample size per chain = 100
1. Empirical mean and standard deviation for each variable,
plus standard error of the mean:
Mean SD Naive SE Time-series SE
sd 2.892e+05 1.971e+03 1.394e+02 2.051e+02
coef -8.384e-02 1.179e-01 8.339e-03 1.070e-02
shape 2.769e-01 2.622e-03 1.854e-04 5.651e-04
rate 2.110e-02 2.239e-04 1.584e-05 4.199e-05
sd_var1 3.560e+02 2.555e+00 1.807e-01 2.480e-01
sd_var2 3.645e+02 2.644e+00 1.870e-01 2.847e-01
2. Quantiles for each variable:
2.5% 25% 50% 75% 97.5%
sd 2.855e+05 2.878e+05 2.894e+05 2.906e+05 2.927e+05
coef -2.332e-01 -1.983e-01 -8.164e-02 3.274e-02 5.738e-02
shape 2.710e-01 2.752e-01 2.773e-01 2.788e-01 2.811e-01
rate 2.068e-02 2.094e-02 2.114e-02 2.127e-02 2.152e-02
sd_var1 3.516e+02 3.540e+02 3.559e+02 3.580e+02 3.607e+02
sd_var2 3.597e+02 3.627e+02 3.646e+02 3.666e+02 3.692e+02
Thank you!