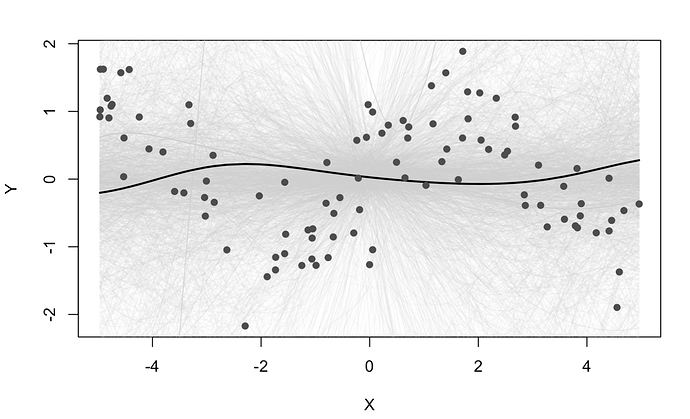
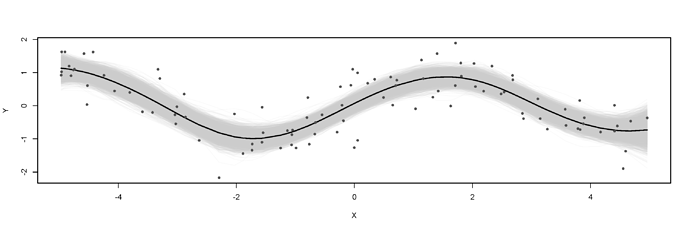
some people have requested RFF greta code - im not sure it works as i wrote it a while back and never tested it, but putting it here incase anyone is curious
Y=
X=
K=500 # number features
omega<-t(as.matrix(rnorm(K)))
library(greta)
tau<-exponential(1) # ridge penalty
bw<-gamma(1,1) # length scale
sigma<-gamma(1,1) # noise
W <- normal(0, 1, dim = c(2K, 1))tau
feat<-X%%(omegabw)
rff<-sqrt(1/K)cbind(sin(feat),cos(feat))
mu <- rff %% W
distribution(Y) <- normal(mu, sigma)
model <- model(W, tau, bw)
draws <- mcmc(model, n_samples = 1000,control = list(Lmin = 10, Lmax = 20, epsilon = 1e-5))