Hi @lionel68!
Thanks so much for sharing this,
I just wanted to post my reprex of your results - I’ll check in with Nick G about this, I haven’t done spatial modelling for a little while, but thanks so much for sharing this! 
library(rstan)
#> Loading required package: StanHeaders
#> Loading required package: ggplot2
#> rstan (Version 2.21.2, GitRev: 2e1f913d3ca3)
#> For execution on a local, multicore CPU with excess RAM we recommend calling
#> options(mc.cores = parallel::detectCores()).
#> To avoid recompilation of unchanged Stan programs, we recommend calling
#> rstan_options(auto_write = TRUE)
library(INLA)
#> Loading required package: Matrix
#> Loading required package: foreach
#> Loading required package: parallel
#> Loading required package: sp
#> This is INLA_21.05.02 built 2021-05-03 11:17:14 UTC.
#> - See www.r-inla.org/contact-us for how to get help.
#> - To enable PARDISO sparse library; see inla.pardiso()
library(RandomFields)
#> Loading required package: RandomFieldsUtils
#>
#> Attaching package: 'RandomFields'
#> The following object is masked from 'package:RandomFieldsUtils':
#>
#> RFoptions
library(greta)
#>
#> Attaching package: 'greta'
#> The following object is masked from 'package:INLA':
#>
#> f
#> The following objects are masked from 'package:Matrix':
#>
#> chol2inv, colMeans, colSums, cov2cor, diag, rowMeans, rowSums
#> The following objects are masked from 'package:stats':
#>
#> binomial, cov2cor, poisson
#> The following objects are masked from 'package:base':
#>
#> %*%, apply, backsolve, beta, chol2inv, colMeans, colSums, diag,
#> eigen, forwardsolve, gamma, identity, rowMeans, rowSums, sweep,
#> tapply
dat <- data.frame(x = runif(100),
y = runif(100))
spat_field <- raster::raster(RFsimulate(RMmatern(nu=1, var = 1, scale = 0.1),
x = seq(0, 1, length.out = 100),
y = seq(0, 1, length.out = 100),
spConform = FALSE))
dat$resp <- rnorm(100, mean = 1 + raster::extract(spat_field, dat), sd = 1)
bnd <- inla.mesh.segment(matrix(c(0, 0, 1, 0, 1, 1, 0, 1), 4, 2, byrow = TRUE))
# a coarse mesh
mesh <- inla.mesh.2d(max.edge = 0.2,
offset = 0,
boundary = bnd)
# derive the FEM matrices
fem <- inla.mesh.fem(mesh)
# put the matrices in one object
G <- array(c(as(fem$c1, "matrix"),
as(fem$g1, "matrix"),
as(fem$g2, "matrix")),
dim = c(mesh$n, mesh$n, 3))
G <- aperm(G, c(3, 1, 2))
# derive the projection matrix
A <- inla.spde.make.A(mesh, loc = as.matrix(dat[,c("x", "y")]))
A <- as(A, "matrix")
y <- dat$resp
# the model parameters
mu <- normal(0, 1)
#> ℹ Initialising python and checking dependencies, this may take a moment.
#> ✓ Initialising python and checking dependencies ... done!
#>
kappa <- lognormal(0, 1) # variance of spatial effect
tau <- gamma(0.1, 0.1) # scale of spatial effect
sigma <- lognormal(0, 1)
z <- normal(0, 1, dim = mesh$n) # std-normal variable for non-centered parametrization
# the precision matrix of the spatial effect
S <- (tau ** 2) * (G[1,,] * kappa ** 4 + G[2,,] * 2 * kappa ** 2 + G[3,,])
# drawing the spatial effect coefficient
beta <- backsolve(chol(S), z)
# the linear predictor
linpred <- mu + A %*% beta
distribution(y) <- normal(linpred, sigma)
# fitting the model
m_g <- model(mu, sigma, tau, kappa)
m_draw <- mcmc(m_g, one_by_one = TRUE)
#> running 4 chains simultaneously on up to 8 cores
#> warmup 0/1000 \| eta: ?s warmup == 50/1000 \| eta: 2m warmup ==== 100/1000 \| eta: 1m warmup ====== 150/1000 \| eta: 1m \| 5% bad warmup ======== 200/1000 \| eta: 1m \| 4% bad warmup ========== 250/1000 \| eta: 43s \| 4% bad warmup =========== 300/1000 \| eta: 38s \| 5% bad warmup ============= 350/1000 \| eta: 35s \| 5% bad warmup =============== 400/1000 \| eta: 31s \| 5% bad warmup ================= 450/1000 \| eta: 28s \| 7% bad warmup =================== 500/1000 \| eta: 25s \| 6% bad warmup ===================== 550/1000 \| eta: 22s \| 6% bad warmup ======================= 600/1000 \| eta: 19s \| 6% bad warmup ========================= 650/1000 \| eta: 17s \| 6% bad warmup =========================== 700/1000 \| eta: 14s \| 5% bad warmup ============================ 750/1000 \| eta: 12s \| 5% bad warmup ============================== 800/1000 \| eta: 9s \| 5% bad warmup ================================ 850/1000 \| eta: 7s \| 5% bad warmup ================================== 900/1000 \| eta: 4s \| 5% bad warmup ==================================== 950/1000 \| eta: 2s \| 4% bad warmup ====================================== 1000/1000 \| eta: 0s \| 4% bad
#> sampling 0/1000 \| eta: ?s sampling == 50/1000 \| eta: 34s sampling ==== 100/1000 \| eta: 32s sampling ====== 150/1000 \| eta: 30s sampling ======== 200/1000 \| eta: 28s sampling ========== 250/1000 \| eta: 26s sampling =========== 300/1000 \| eta: 24s sampling ============= 350/1000 \| eta: 23s sampling =============== 400/1000 \| eta: 21s \| \<1% bad sampling ================= 450/1000 \| eta: 20s \| \<1% bad sampling =================== 500/1000 \| eta: 18s \| \<1% bad sampling ===================== 550/1000 \| eta: 17s \| \<1% bad sampling ======================= 600/1000 \| eta: 15s \| 1% bad sampling ========================= 650/1000 \| eta: 13s \| 2% bad sampling =========================== 700/1000 \| eta: 11s \| 2% bad sampling ============================ 750/1000 \| eta: 9s \| 2% bad sampling ============================== 800/1000 \| eta: 8s \| 1% bad sampling ================================ 850/1000 \| eta: 6s \| 1% bad sampling ================================== 900/1000 \| eta: 4s \| 1% bad sampling ==================================== 950/1000 \| eta: 2s \| 1% bad sampling ====================================== 1000/1000 \| eta: 0s \| 1% bad
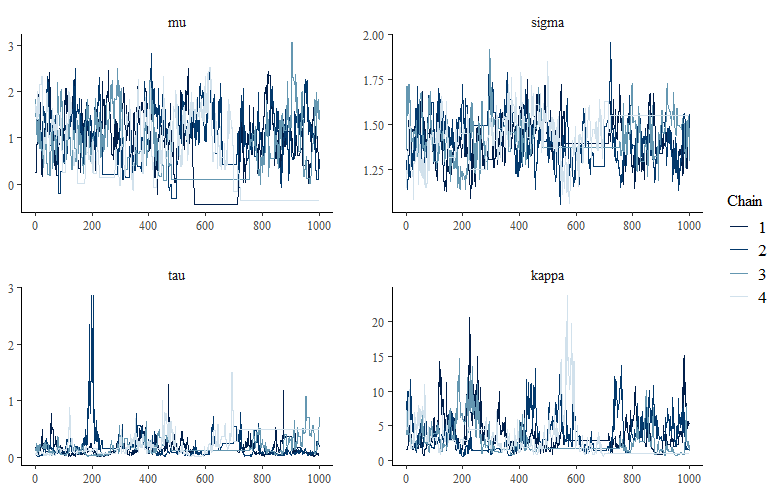
bayesplot::mcmc_trace(m_draw)
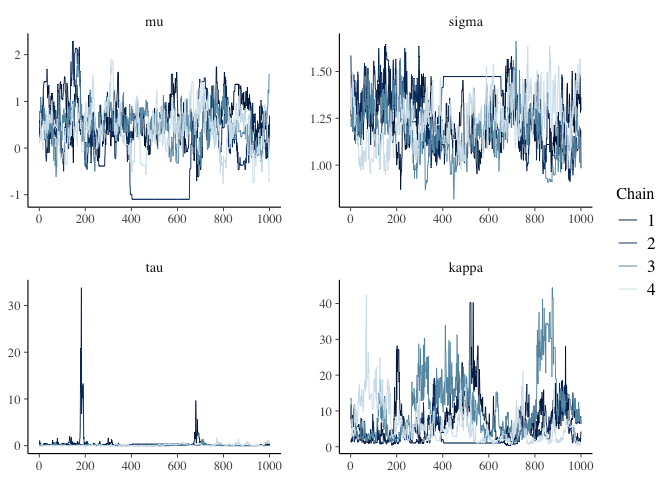
coda::gelman.diag(m_draw)
#> Potential scale reduction factors:
#>
#> Point est. Upper C.I.
#> mu 1.25 1.70
#> sigma 1.13 1.37
#> tau 1.22 1.56
#> kappa 1.30 1.93
#>
#> Multivariate psrf
#>
#> 1.23
Created on 2021-11-04 by the reprex package (v2.0.1)
Session info
sessioninfo::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.1.1 (2021-08-10)
#> os macOS Big Sur 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_AU.UTF-8
#> ctype en_AU.UTF-8
#> tz Australia/Perth
#> date 2021-11-04
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.1.0)
#> assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.1.0)
#> backports 1.2.1 2020-12-09 [1] CRAN (R 4.1.0)
#> base64enc 0.1-3 2015-07-28 [1] CRAN (R 4.1.0)
#> bayesplot 1.8.1 2021-06-14 [1] CRAN (R 4.1.0)
#> callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
#> cli 3.1.0 2021-10-27 [1] CRAN (R 4.1.1)
#> coda 0.19-4 2020-09-30 [1] CRAN (R 4.1.0)
#> codetools 0.2-18 2020-11-04 [1] CRAN (R 4.1.1)
#> colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.0)
#> crayon 1.4.1 2021-02-08 [1] CRAN (R 4.1.0)
#> curl 4.3.2 2021-06-23 [1] CRAN (R 4.1.0)
#> DBI 1.1.1 2021-01-15 [1] CRAN (R 4.1.0)
#> digest 0.6.28 2021-09-23 [1] CRAN (R 4.1.0)
#> dplyr 1.0.7 2021-06-18 [1] CRAN (R 4.1.0)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
#> evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
#> fansi 0.5.0 2021-05-25 [1] CRAN (R 4.1.0)
#> farver 2.1.0 2021-02-28 [1] CRAN (R 4.1.0)
#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
#> foreach * 1.5.1 2020-10-15 [1] CRAN (R 4.1.0)
#> fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.0)
#> future 1.23.0 2021-10-31 [1] CRAN (R 4.1.1)
#> generics 0.1.1 2021-10-25 [1] CRAN (R 4.1.0)
#> ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.1.0)
#> ggridges 0.5.3 2021-01-08 [1] CRAN (R 4.1.0)
#> globals 0.14.0 2020-11-22 [1] CRAN (R 4.1.0)
#> glue 1.4.2 2020-08-27 [1] CRAN (R 4.1.0)
#> greta * 0.3.1.9012 2021-11-03 [1] local
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.1.0)
#> gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
#> here 1.0.1 2020-12-13 [1] CRAN (R 4.1.0)
#> highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
#> hms 1.1.1 2021-09-26 [1] CRAN (R 4.1.0)
#> htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
#> httr 1.4.2 2020-07-20 [1] CRAN (R 4.1.0)
#> INLA * 21.05.02 2021-05-03 [1] local
#> inline 0.3.19 2021-05-31 [1] CRAN (R 4.1.0)
#> iterators 1.0.13 2020-10-15 [1] CRAN (R 4.1.0)
#> jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
#> knitr 1.36 2021-09-29 [1] CRAN (R 4.1.0)
#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.1.0)
#> lattice 0.20-44 2021-05-02 [1] CRAN (R 4.1.1)
#> lifecycle 1.0.1 2021-09-24 [1] CRAN (R 4.1.0)
#> listenv 0.8.0 2019-12-05 [1] CRAN (R 4.1.0)
#> loo 2.4.1 2020-12-09 [1] CRAN (R 4.1.0)
#> magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
#> Matrix * 1.3-4 2021-06-01 [1] CRAN (R 4.1.1)
#> matrixStats 0.61.0 2021-09-17 [1] CRAN (R 4.1.0)
#> mime 0.12 2021-09-28 [1] CRAN (R 4.1.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
#> parallelly 1.28.1 2021-09-09 [1] CRAN (R 4.1.0)
#> pillar 1.6.4 2021-10-18 [1] CRAN (R 4.1.0)
#> pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
#> plyr 1.8.6 2020-03-03 [1] CRAN (R 4.1.0)
#> png 0.1-7 2013-12-03 [1] CRAN (R 4.1.0)
#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
#> processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
#> progress 1.2.2 2019-05-16 [1] CRAN (R 4.1.0)
#> ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
#> purrr 0.3.4 2020-04-17 [1] CRAN (R 4.1.0)
#> R.cache 0.15.0 2021-04-30 [1] CRAN (R 4.1.0)
#> R.methodsS3 1.8.1 2020-08-26 [1] CRAN (R 4.1.0)
#> R.oo 1.24.0 2020-08-26 [1] CRAN (R 4.1.0)
#> R.utils 2.11.0 2021-09-26 [1] CRAN (R 4.1.0)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
#> RandomFields * 3.3.8 2020-02-06 [1] CRAN (R 4.1.0)
#> RandomFieldsUtils * 0.5.4 2021-09-22 [1] CRAN (R 4.1.0)
#> raster 3.4-13 2021-06-18 [1] CRAN (R 4.1.0)
#> Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.1.0)
#> RcppParallel 5.1.4 2021-05-04 [1] CRAN (R 4.1.0)
#> reprex 2.0.1 2021-08-05 [1] CRAN (R 4.1.0)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.1.0)
#> reticulate 1.22 2021-09-17 [1] CRAN (R 4.1.0)
#> rgdal 1.5-27 2021-09-16 [1] CRAN (R 4.1.0)
#> rlang 0.4.12 2021-10-18 [1] CRAN (R 4.1.0)
#> rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.0)
#> rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
#> rstan * 2.21.2 2020-07-27 [1] CRAN (R 4.1.0)
#> rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
#> scales 1.1.1 2020-05-11 [1] CRAN (R 4.1.0)
#> sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.1.0)
#> sp * 1.4-5 2021-01-10 [1] CRAN (R 4.1.0)
#> StanHeaders * 2.21.0-7 2020-12-17 [1] CRAN (R 4.1.0)
#> stringi 1.7.5 2021-10-04 [1] CRAN (R 4.1.0)
#> stringr 1.4.0 2019-02-10 [1] CRAN (R 4.1.0)
#> styler 1.6.2 2021-09-23 [1] CRAN (R 4.1.0)
#> tensorflow 2.6.0 2021-08-19 [1] CRAN (R 4.1.0)
#> tfruns 1.5.0 2021-02-26 [1] CRAN (R 4.1.0)
#> tibble 3.1.5 2021-09-30 [1] CRAN (R 4.1.0)
#> tidyselect 1.1.1 2021-04-30 [1] CRAN (R 4.1.0)
#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
#> V8 3.4.2 2021-05-01 [1] CRAN (R 4.1.0)
#> vctrs 0.3.8 2021-04-29 [1] CRAN (R 4.1.0)
#> whisker 0.4 2019-08-28 [1] CRAN (R 4.1.0)
#> withr 2.4.2 2021-04-18 [1] CRAN (R 4.1.0)
#> xfun 0.26 2021-09-14 [1] CRAN (R 4.1.0)
#> xml2 1.3.2 2020-04-23 [1] CRAN (R 4.1.0)
#> yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.1/Resources/library